ņł▓(forest)ņØ┤ ņ׳Ļ│Ā, ņł▓ņåŹņŚÉļŖö ļéśļ¼┤(tree)ļōżņØ┤ ņ׳ļŗż. ņŚ¼ĻĖ░ņä£ ļéśļ¼┤ļŖö ņØśņé¼Ļ▓░ņĀĢĒŖĖļ”¼ļŗż. input ļŹ░ņØ┤Ēä░ļŖö randomņØ┤Ļ│Ā, ņł▓ļÅä randomņØ┤ļŗż. ņł▓ņŚÉ ņ׳ļŖö ļéśļ¼┤ļōżņŚÉĻ▓ī random inputņØä ĒĢ┤ņä£ Ļ░ü ļéśļ¼┤ļōżņØ┤ ļ▒ēņ¢┤ļé┤ļŖö Ļ▓░Ļ│╝ļź╝ voting(ļŗżņłśĻ▓░ņØś ņøÉņ╣Ö)ĒĢ┤ņä£ ļČäļźśĒĢ£ļŗż. ļīĆņÜ®ļ¤ē ļŹ░ņØ┤Ēä░ņŚÉ ĒÜ©Ļ│╝ņĀüņ£╝ļĪ£ ņŗżĒ¢ēļÉśĻ│Ā, ļ¦ÄņØĆ ļ│Ćņłśļź╝ ņØ┤ņÜ®ĒĢ┤ļÅä ļ│Ćņłś ņĀ£Ļ▒░ ņŚåņØ┤ ņŗżĒ¢ēļÉśņ¢┤ ņĀĢĒÖĢļÅäĻ░Ć ļåÆņØĆ ĒÄĖņØ┤ļŗż. unbalancedļÉ£ classņØś ļ¬©ņ¦æļŗ©ņŚÉ ļīĆĒĢ┤ ņל ļ¦×ļŖöļŗż. -- RņØä ņØ┤ņÜ®ĒĢ£ ļ╣ģļŹ░ņØ┤Ēä░ ļČäņäØ, Ļ╣ĆĻ▓ĮĒā£ ņ░ĖĻ│Ā
1 ņśłņĀ£ ļŹ░ņØ┤Ēä░ #
![[http]](/moniwiki/imgs/http.png) EXCELņŚÉ ņØśĒĢ£ ņĪ░ņé¼ļ░®ļ▓Ģ ļ░Å ĒåĄĻ│äļČäņäØ(http://www.kyobobook.co.kr/product/detailViewKor.laf?ejkGb=KOR&mallGb=KOR&barcode=9788983257000&orderClick=LAG&Kc=SETLBkserp11_15)
EXCELņŚÉ ņØśĒĢ£ ņĪ░ņé¼ļ░®ļ▓Ģ ļ░Å ĒåĄĻ│äļČäņäØ(http://www.kyobobook.co.kr/product/detailViewKor.laf?ejkGb=KOR&mallGb=KOR&barcode=9788983257000&orderClick=LAG&Kc=SETLBkserp11_15)ļź╝ ņØ┤ņÜ®ĒĢ©.
cname <- c("ID", "ĻĄ¼ļ¦żļĖīļ×£ļō£", "ņŚ░ļĀ╣","ņäĖļīĆņŚ░ņłśņ×ģ", "ņäĖļīĆņé¼ļ×īņłś", "ļ░®ļ¼Ėļ╣łļÅä", "Ļ▒░ņŻ╝ļģäņłś")
x = read.table("c:\\data\\disc.txt", col.names = cname)
head(x)
 disc.txt
disc.txt
> head(x)
ID ĻĄ¼ļ¦żļĖīļ×£ļō£ ņŚ░ļĀ╣ ņäĖļīĆņŚ░ņłśņ×ģ ņäĖļīĆņé¼ļ×īņłś ļ░®ļ¼Ėļ╣łļÅä Ļ▒░ņŻ╝ļģäņłś
1 1 A 48 9000 4 5 6
2 2 A 58 8000 6 4 20
3 3 A 52 7000 6 4 12
4 4 A 63 7000 6 4 15
5 5 A 59 8000 4 6 6
6 6 A 38 11000 5 4 10
>
2 ļ×£ļŹżĒżļĀłņŖżĒŖĖ #
tree <- randomForest(ĻĄ¼ļ¦żļĖīļ×£ļō£ ~ ņŚ░ļĀ╣ + ņäĖļīĆņŚ░ņłśņ×ģ + ņäĖļīĆņé¼ļ×īņłś + ļ░®ļ¼Ėļ╣łļÅä + Ļ▒░ņŻ╝ļģäņłś, data=x)
print(tree) # view results
importance(tree)
Ļ▓░Ļ│╝
> print(tree) # view results
Call:
randomForest(formula = ĻĄ¼ļ¦żļĖīļ×£ļō£ ~ ņŚ░ļĀ╣ + ņäĖļīĆņŚ░ņłśņ×ģ + ņäĖļīĆņé¼ļ×īņłś + ļ░®ļ¼Ėļ╣łļÅä + Ļ▒░ņŻ╝ļģäņłś, data = x)
Type of random forest: classification
Number of trees: 500
No. of variables tried at each split: 2
OOB estimate of error rate: 5%
Confusion matrix:
A B class.error
A 10 0 0.0
B 1 9 0.1
> importance(tree)
MeanDecreaseGini
ņŚ░ļĀ╣ 2.624620
ņäĖļīĆņŚ░ņłśņ×ģ 1.815804
ņäĖļīĆņé¼ļ×īņłś 1.263035
ļ░®ļ¼Ėļ╣łļÅä 1.196015
Ļ▒░ņŻ╝ļģäņłś 2.576659
>
ĻĄ¼ļ¦żļĖīļ×£ļō£ļź╝ Ļ▓░ņĀĢĒĢśļŖö ļ│ĆņłśņØś ņżæņÜöļÅäļŖö ņŚ░ļĀ╣ > Ļ▒░ņŻ╝ļģäņłś > ņäĖļīĆņŚ░ņłśņ×ģ > ņäĖļīĆņé¼ļ×īņłś > ļ░®ļ¼Ėļ╣łļÅä ņł£ņØ┤ļŗż.
rf <- randomForest(factor(t3)~diff_cnt+diff_time, data=x6, type="classification", importance=TRUE,na.action=na.omit)
pred <- predict(rf, newdata=test)
table(pred, test$t3)
data(iris)
set.seed(111)
ind <- sample(2, nrow(iris), replace = TRUE, prob=c(0.8, 0.2))
iris.rf <- randomForest(Species ~ ., data=iris[ind == 1,])
iris.pred <- predict(iris.rf, iris[ind == 2,])
table(observed = iris[ind==2, "Species"], predicted = iris.pred)
ĒöīļĪ£Ēīģ..
install.packages("rpart")
library("rpart")
cf <- cforest(Species ~ ., data = iris)
pt <- party:::prettytree(cf@ensemble[[1]], names(cf@data@get("input")))
pt
nt <- new("BinaryTree")
nt@tree <- pt
nt@data <- cf@data
nt@responses <- cf@responses
nt
plot(nt)
install.packages("tree")
library(tree)
tr <- tree(Species ~ ., data=iris)
tr
3 ļ│Ćņłś ņżæņÜöļÅäņÖĆ Gini impurity #
Gini impurity
ņ¦Ćļŗł ņ¦Ćņłś(Gini Index)ļŖö ļČłņł£ļÅä(impurity)ļź╝ ņĖĪņĀĢĒĢśļŖö ĒĢśļéśņØś ņ¦ĆņłśņØ┤ļŗż. ņ×äņØśņØś ĒĢ£ Ļ░£ņ▓┤Ļ░Ć ļ¬®Ēæ£ļ│ĆņłśņØś iļ▓łņ¦Ė ļ▓öņŻ╝ļĪ£ļČĆĒä░ ņČöņČ£ļÉśņŚłĻ│Ā, ĻĘĖ Ļ░£ņ▓┤ļź╝ ļ¬®Ēæ£ļ│ĆņłśņØś jļ▓łņ¦Ė ļ▓öņŻ╝ņŚÉ ņåŹĒĢ£ļŗżĻ│Ā ņśżļČäļźś(misclassification)ĒĢĀ ĒÖĢļźĀņØĆ P(i)P(j)Ļ░Ć ļÉ£ļŗż. ņŚ¼ĻĖ░ņŚÉņä£ P(i)ļŖö Ļ░ü ļ¦łļööņŚÉņä£ ĒĢ£ Ļ░£ņ▓┤Ļ░Ć ļ¬®Ēæ£ļ│ĆņłśņØś Iļ▓łņ¦Ė ļ▓öņŻ╝ņŚÉ ņåŹĒĢĀ ĒÖĢļźĀņØ┤ļŗż. ņØ┤ļ¤¼ĒĢ£ ņśżļČäļźś ĒÖĢļźĀņØĆ ļ¬©ļæÉ ļŹöĒĢśņŚ¼
ļ│Ćņłś ņżæņÜöļÅä
imp <- data.frame(importance(model))
imp[order(imp$MeanDecreaseGini, decreasing=T),]
varImpPlot(model)
Ļ▓░Ļ│╝ļŖö ņĀĢĒÖĢļÅä Ļ┤ĆņĀÉņŚÉņä£ņØś ņżæņÜöļÅäņÖĆ ņ¦Ćļŗł ļČłņł£ļÅä Ļ┤ĆņĀÉņŚÉņä£ ņżæņÜöļÅä 2Ļ░Ćņ¦ĆļĪ£ plotting ļÉ£ļŗż.
4 RRF #
Regularized Random Forest
install.packages("RRF")
library("RRF")
model <- RRF(factor(is_out) ~ ., data=training, type="classification", importance=TRUE)
pred <- predict(model, newdata=test2)
confusionMatrix(pred, test2$is_out)
![[http]](/moniwiki/imgs/http.png) EXCELņŚÉ ņØśĒĢ£ ņĪ░ņé¼ļ░®ļ▓Ģ ļ░Å ĒåĄĻ│äļČäņäØ(http://www.kyobobook.co.kr/product/detailViewKor.laf?ejkGb=KOR&mallGb=KOR&barcode=9788983257000&orderClick=LAG&Kc=SETLBkserp11_15)ļź╝ ņØ┤ņÜ®ĒĢ©.
EXCELņŚÉ ņØśĒĢ£ ņĪ░ņé¼ļ░®ļ▓Ģ ļ░Å ĒåĄĻ│äļČäņäØ(http://www.kyobobook.co.kr/product/detailViewKor.laf?ejkGb=KOR&mallGb=KOR&barcode=9788983257000&orderClick=LAG&Kc=SETLBkserp11_15)ļź╝ ņØ┤ņÜ®ĒĢ©. disc.txt
disc.txt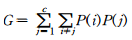
![[https]](/moniwiki/imgs/https.png)