кё°ліём ҒмңјлЎң м•„лһҳмІҳлҹј н•ңлӢӨ.
cname <- c("x1", "x2", "x3","x4", "x5", "x6", "y")
german = read.table("c:\\data\\german1.txt", col.names = cname)
options(contrasts = c("contr.treatment", "contr.poly"))
german.object <- glm(y~.,family=binomial, data=german)
summary(german.object)
p <- predict(german.object, german, type="response")
german <- cbind(german, p)
head(german)
pлҠ” y=1мқј нҷ•лҘ мқҙлӢӨ. көҗм ңм—җм„ңлҠ” y=1мқҙ л¶Ҳлҹүкі к°қмқҙлӢӨ.
> head(german)
x1 x2 x3 x4 x5 x6 y p
1 A11 6 A34 4 67 1 0 0.18170622
2 A12 48 A32 2 22 1 1 0.63312134
3 A14 12 A34 3 49 2 0 0.04812842
4 A11 42 A32 4 45 2 0 0.64393801
5 A11 24 A33 4 53 2 1 0.47102819
6 A14 36 A32 4 35 2 0 0.20127115
4 ліҖмҲҳм„ нғқлІ• #
м „м§„м„ нғқ, ліҖмҲҳмҶҢкұ°, л‘ҳлӢӨ
cname <- c("x1", "x2", "x3","x4", "x5", "x6", "y")
german = read.table("c:\\data\\german1.txt", col.names = cname)
#мғҒмҲҳн•ӯл§Ң нҸ¬н•Ён•ң мҙҲкё°лӘЁнҳ•
g.object <- glm(y~1, family=binomial, data=german)
#м „м§„м„ нғқлІ•
s.list <- list(upper = update(g.object, ~. +x1+x2+x3+x4+x5+x6))
step(g.object, scope=s.list, direction="forward")
#ліҖмҲҳмҶҢкұ°лІ•
#s.list <- list(lower = update(g.object, ~. -x1-x2-x3-x4-x5-x6))
#step(g.object, scope=s.list, direction="backward")
#л‘ҳлӢӨ
#s.list <- list(upper = update(g.object, ~. +x1+x2+x3+x4+x5+x6), lower = update(g.object, ~. -x1-x2-x3-x4-x5-x6))
#step(g.object, scope=s.list, direction="both")
кІ°кіј
> step(g.object, scope=s.list, direction="forward")
Start: AIC=1223.73
y ~ 1
Df Deviance AIC
+ x1 3 1090.4 1098.4
+ x3 4 1161.3 1171.3
+ x2 1 1177.1 1181.1
+ x5 1 1213.1 1217.1
<none> 1221.7 1223.7
+ x6 1 1221.7 1225.7
+ x4 1 1221.7 1225.7
Step: AIC=1098.39
y ~ x1
Df Deviance AIC
+ x2 1 1051.9 1061.9
+ x3 4 1053.2 1069.2
+ x5 1 1085.0 1095.0
<none> 1090.4 1098.4
+ x4 1 1090.2 1100.2
+ x6 1 1090.3 1100.3
Step: AIC=1061.9
y ~ x1 + x2
Df Deviance AIC
+ x3 4 1022.6 1040.6
+ x5 1 1046.7 1058.7
<none> 1051.9 1061.9
+ x4 1 1051.5 1063.5
+ x6 1 1051.9 1063.9
Step: AIC=1040.58
y ~ x1 + x2 + x3
Df Deviance AIC
+ x5 1 1019.3 1039.3
<none> 1022.6 1040.6
+ x6 1 1022.5 1042.5
+ x4 1 1022.5 1042.5
Step: AIC=1039.27
y ~ x1 + x2 + x3 + x5
Df Deviance AIC
<none> 1019.3 1039.3
+ x4 1 1019.2 1041.2
+ x6 1 1019.3 1041.3
Call: glm(formula = y ~ x1 + x2 + x3 + x5, family = binomial, data = german)
Coefficients:
(Intercept) x1A12 x1A13 x1A14 x2
0.66510 -0.53186 -1.07608 -1.89880 0.03434
x3A31 x3A32 x3A33 x3A34 x5
-0.09915 -0.94696 -0.93433 -1.52607 -0.01277
Degrees of Freedom: 999 Total (i.e. Null); 990 Residual
Null Deviance: 1222
Residual Deviance: 1019 AIC: 1039
>
мөңмў…м ҒмңјлЎң glm(formula = y ~ x1 + x2 + x3 + x5, family = binomial, data = german) к°Җ м„ нғқлҗҳм—ҲлӢӨ. AICк°Җ к°ҖмһҘ м ҒмқҖ лӘЁлҚёмқҙ м„ нғқлҗңлӢӨ.
м°ёкі : AIC (
![[http]](/moniwiki/imgs/http.png) м¶ңмІҳ(http://blog.naver.com/pinokio129?Redirect=Log&logNo=80018702687)
м¶ңмІҳ(http://blog.naver.com/pinokio129?Redirect=Log&logNo=80018702687))
AIC нҶөкі„лҹү
мөңм Ғ лӘЁнҳ•мқ„ кІ°м •н•ҳлҠ” кІҪмҡ° лӘЁнҳ•мқҳ м„ӨлӘ…лҠҘл Ҙкіј лӘЁнҳ•мқҳ м Ҳм•Ҫм„ұмқ„ кі л Өн•ҙм•ј н•Ё.
лӘЁнҳ•мқҳ м„ӨлӘ…лҠҘл ҘмқҖ мҡ°лҸ„(likelihood)лЎңм„ң мёЎм •лҗ мҲҳ мһҲмңјл©°,
лӘЁнҳ•мқҳ м Ҳм•Ҫм„ұмқҖ ліөмһЎн•ң лӘЁнҳ• ліҙлӢӨлҠ” к°„кІ°н•ң лӘЁнҳ•мқҙ м„ нҳёлҗҳм–ҙм•ј н•ңлӢӨлҠ” кІғмңјлЎң
м¶”м • кі„мҲҳмқҳ ж•ёк°Җ л§ҺмқҖ кІғм—җ лҢҖн•ҙ лІҢм№ҷмқ„ к°Җн•ЁмңјлЎңмҚЁ кі л Өлҗ мҲҳ мһҲлӢӨ.
к·ёлҹ¬лӮҳ лӘЁнҳ•мқҳ м„ӨлӘ…л Ҙкіј м Ҳм•Ҫм„ұмқҖ мғҒ충лҗҳлҠ”(trade-off)кё°мӨҖмңјлЎңм„ң,
мҷ•мҷ• ліөмһЎн•ң лӘЁнҳ•мқҙ м„ӨлӘ…л Ҙмқҙ нҒ°кІғмқ„ л°ңкІ¬ н• мҲҳ мһҲлӢӨ.
л”°лқјм„ң л‘җ нҢҗлӢЁкё°мӨҖмқ„ н•Ёк»ҳ кі л Өн•ң м„ нғқк·ңмӨҖмқҙ н•„мҡ”н•ҳкІҢ лҗңлӢӨ.
к·ёкІғмқҙ л°”лЎң м •ліҙкё°мӨҖ м ‘к·јлІ•мңјлЎң Akailkeм—җ мқҳн•ҳм—¬ к°ңл°ңлҗң
Akaike Information Criterion(AIC)мқҙлӢӨ.
AIC = -2 log L +2n
м—¬кё°м„ң 'n': нҢҢлқјлҜён„° мҲҳлҘј лӮҳнғҖлӮҙл©°, 'log L'мқҖ лҢҖмҲҳмҡ°лҸ„(log-likelihood).
вҖ» Akailkeм—җ мқҳн•ҳл©ҙ AICнҶөкі„лҹүмқҳ к°’мқҙ мөңмҶҢк°Җ лҗҳлҠ” лӘЁнҳ•мқ„ мұ„нғқн•ҳм—¬м•ј лҗңлӢӨ
D(Deviance)лҠ” мқјл°ҳнҷ”лҗң м„ нҳ• лӘЁнҳ•м—җм„ң лӘЁнҳ•мқҳ м Ғн•©лҸ„ кІҖмҰқ нҶөлҹүмқҙлӢӨ.
cname <- c("x1", "x2", "x3","x4", "x5", "x6", "y")
german = read.table("c:\\data\\german1.txt", col.names = cname)
options(contrasts = c("contr.treatment", "contr.poly"))
german.object <- glm(formula = y ~ x1 + x2 + x3 + x5, family = binomial, data = german)
summary(german.object)
кІ°кіј
Call:
glm(formula = y ~ x1 + x2 + x3 + x5, family = binomial, data = german)
Deviance Residuals:
Min 1Q Median 3Q Max
-1.7997 -0.8007 -0.4747 0.9283 2.4241
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 0.665099 0.476311 1.396 0.162607
x1A12 -0.531864 0.185402 -2.869 0.004121 **
x1A13 -1.076082 0.336956 -3.194 0.001405 **
x1A14 -1.898800 0.206314 -9.203 < 2e-16 ***
x2 0.034342 0.006329 5.426 5.76e-08 ***
x3A31 -0.099153 0.474942 -0.209 0.834629
x3A32 -0.946961 0.370644 -2.555 0.010622 *
x3A33 -0.934327 0.433303 -2.156 0.031061 *
x3A34 -1.526069 0.393754 -3.876 0.000106 ***
x5 -0.012765 0.007091 -1.800 0.071821 .
---
Signif. codes: 0 вҖҳ***вҖҷ 0.001 вҖҳ**вҖҷ 0.01 вҖҳ*вҖҷ 0.05 вҖҳ.вҖҷ 0.1 вҖҳ вҖҷ 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 1221.7 on 999 degrees of freedom
Residual deviance: 1019.3 on 990 degrees of freedom
AIC: 1039.3
Number of Fisher Scoring iterations: 4
мҲ«мһҗнҳ• ліҖмҲҳмқё кІҪмҡ° 분нҸ¬лҘј ліҙкі нӣ„ліҙлҘј м„ нғқн• мҲҳ мһҲлӢӨ. (YлӢҳмқҙ м•Ңл ӨмӨҖкұ°лӢӨ)
# r: кІ°кіјset
# rм—җм„ң мҲ«мһҗнҳ• мһҗлЈҢл§Ң м„ лі„н•ҙм„ң plotting
# columnмқҙ мҲ«мһҗнҳ•мқём§Җ T/F м ҖмһҘ
training <- sqlQuery(conn, "select
cnt x1
, logkind x4
, genre x5
, diff_cnt x6
, diff_time x8
, gamecd x9
, reg_hh x10
, case when t <= 2 then 1 else 0 end y
from temp.dbo.out_data01
")
#install.packages("reshape")
#install.packages("ggplot2")
library("reshape")
library("ggplot2")
number_cols <- sapply(training, is.numeric)
r.lng <- melt(training[,number_cols], id="y") # id: мў…мҶҚліҖмҲҳлӘ….
p <- ggplot(aes(x=value, group=y, colour=factor(y)), data=r.lng)
p + geom_density() + facet_wrap(~variable,scale="free")
кІ°кіјлҠ” лӢӨмқҢ к·ёлҹјкіј к°ҷлӢӨ.
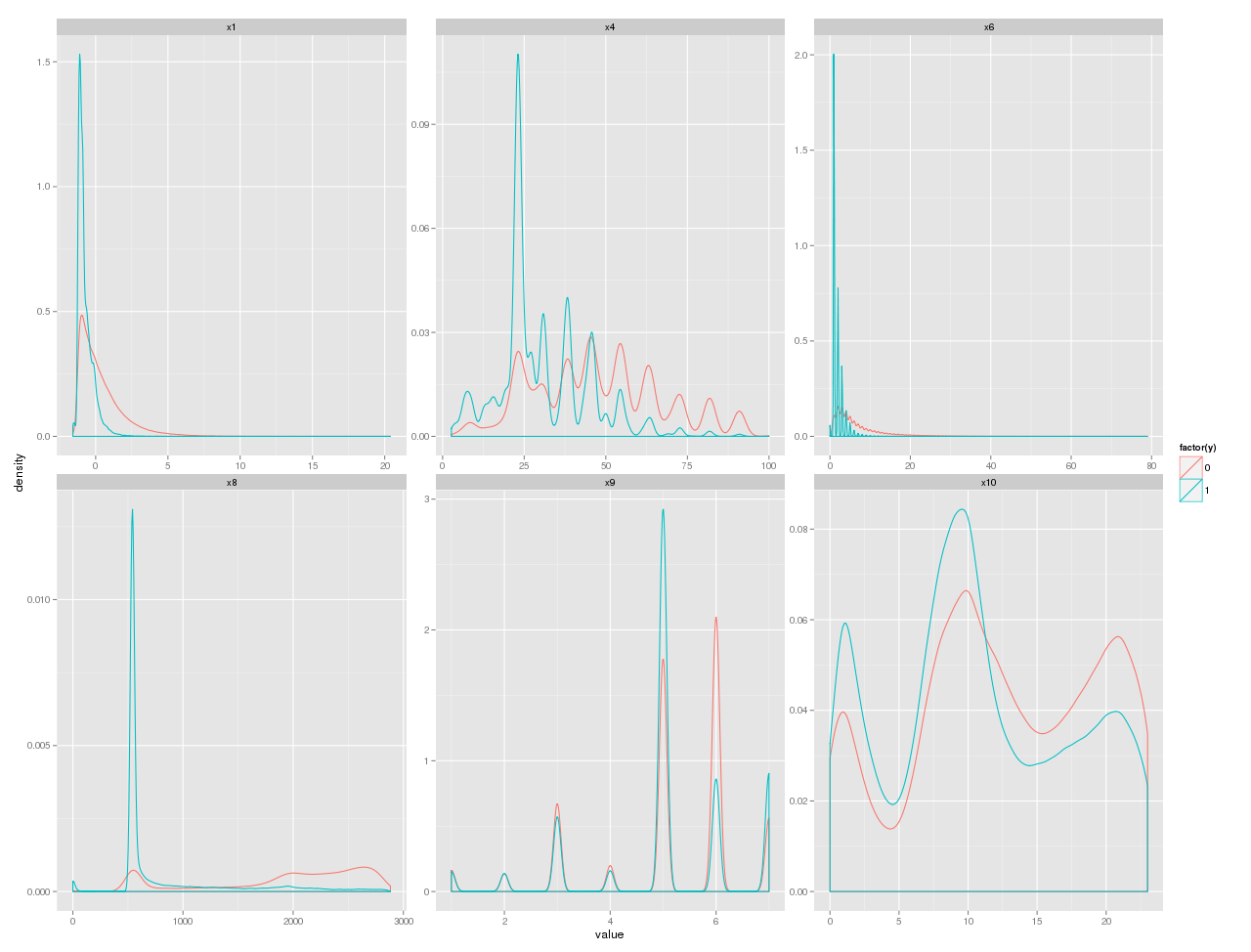
м—¬кё°м—җм„ңлҠ” к·ёлҰјмқ„ ліҙкі x6, x8мқҙ м„ нғқлҗҳм—ҲлӢӨ.
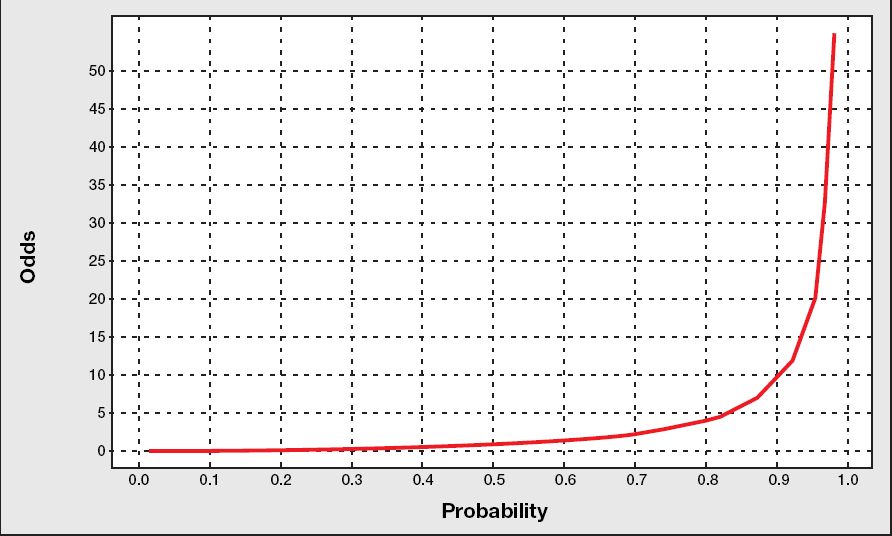
5 oddsмҷҖ logit #
мҠ№мӮ°(к·ёлҰј м¶ңмІҳ ->
![[http]](/moniwiki/imgs/http.png) м—¬кё°(http://www.bisolutions.us/Credit-Risk-Evaluation-of-Online-Personal-Loan-Applicants-A-Data-Mining-Approach.php)
м—¬кё°(http://www.bisolutions.us/Credit-Risk-Evaluation-of-Online-Personal-Loan-Applicants-A-Data-Mining-Approach.php))
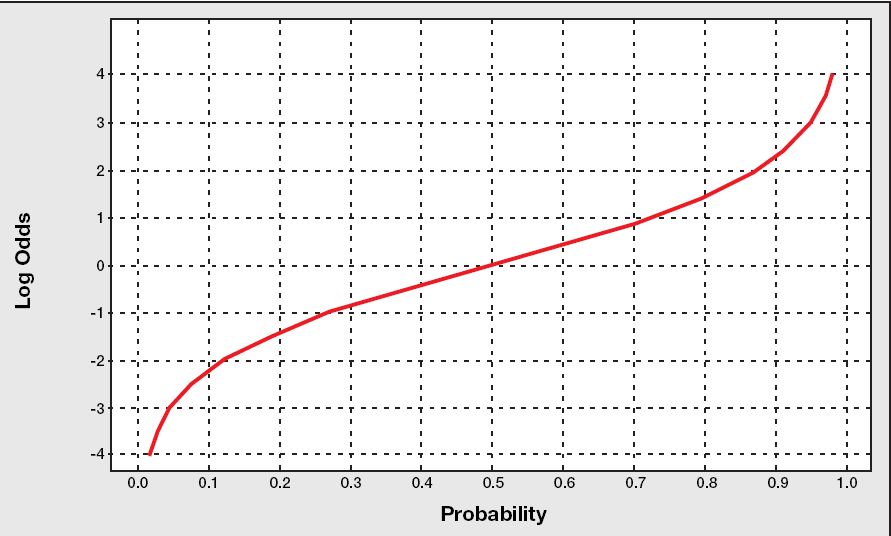
лЎң짓(к·ёлҰј м¶ңмІҳ ->
![[http]](/moniwiki/imgs/http.png) м—¬кё°(http://www.bisolutions.us/Credit-Risk-Evaluation-of-Online-Personal-Loan-Applicants-A-Data-Mining-Approach.php)
м—¬кё°(http://www.bisolutions.us/Credit-Risk-Evaluation-of-Online-Personal-Loan-Applicants-A-Data-Mining-Approach.php)) = logмҷҖ oddsлҘј н•©м№ңл§җ = log(1/(1-p))
pred <- predict(german.object, german, type="response")
logodds <- predict(german.object, german)
german <- cbind(german, pred) #нҷ•лҘ
german <- cbind(german, logodds) #мҳӨмҰҲ, odds
head(german)
кІ°кіј
> head(german)
x1 x2 x3 x4 x5 x6 y pred logodds
1 A11 6 A34 4 67 1 0 0.18091034 -1.5101920
2 A12 48 A32 2 22 1 1 0.63503244 0.5538676
3 A14 12 A34 3 49 2 0 0.04865313 -2.9731627
4 A11 42 A32 4 45 2 0 0.64246423 0.5860757
5 A11 24 A33 4 53 2 1 0.46964395 -0.1215737
6 A14 36 A32 4 35 2 0 0.19922819 -1.3911252
2лІҲм§ё RowлҠ”
| x1 | x2 | x3 | x4 | x5 | x6 | y |
| A12 | 48 | A32 | 2 | 22 | 1 | 1 |
мқҙкі , нҡҢк·ҖмӢқмқҙ лӢӨмқҢкіј к°ҷмңјлҜҖлЎң
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 0.665099 0.476311 1.396 0.162607
x1A12 -0.531864 0.185402 -2.869 0.004121 **
x1A13 -1.076082 0.336956 -3.194 0.001405 **
x1A14 -1.898800 0.206314 -9.203 < 2e-16 ***
x2 0.034342 0.006329 5.426 5.76e-08 ***
x3A31 -0.099153 0.474942 -0.209 0.834629
x3A32 -0.946961 0.370644 -2.555 0.010622 *
x3A33 -0.934327 0.433303 -2.156 0.031061 *
x3A34 -1.526069 0.393754 -3.876 0.000106 ***
x5 -0.012765 0.007091 -1.800 0.071821 .
лӢӨмқҢкіј к°ҷмқҙ кі„мӮ°лҗңлӢӨ.
0.665099 -0.531864 + 48 * 0.034342 - 0.946961 -0.012765 * 22 = 0.5538676
нҷ•лҘ мқҖ..
x <- 0.665099 -0.531864 + 48 * 0.034342 - 0.946961 -0.012765 * 22
1 / (1 + exp(-x)) #кІ°кіјк°’ 0.6350307
exp(x) / (1 + exp(x)) #кІ°кіјк°’ 0.6350307, мқҙкІҢ лЎңм§ҖмҠӨнӢұ лӘЁнҳ•
кіј к°ҷмқҙ кі„мӮ°н•ҳл©ҙ лҗңлӢӨ.
8 분лҘҳн‘ң(confusion matrix) #
#T1 <- length(german[german$y==1 & german$pred > 0.5,]$x1)
#T0 <- length(german[german$y==0 & german$pred > 0.5,]$x1)
#F1 <- length(german[german$y==1 & german$pred <= 0.5,]$x1)
#F0 <- length(german[german$y==0 & german$pred <= 0.5,]$x1)
#
install.packages("SDMTools")
library("SDMTools")
confusion.matrix(german$y,german$pred,threshold=0.5)
кІ°кіј
> confusion.matrix(german$y,german$pred,threshold=0.5)
obs
pred 0 1
0 629 177
1 71 123
attr(,"class")
[1] "confusion.matrix"
| | False(мӢӨм ң) | True(мӢӨм ң) |
| False(мҳҲмёЎ) | 629 | 177 |
| True(мҳҲмёЎ) | 71 | 123 |
| н•ӯлӘ© | кі„мӮ° | нҷ•лҘ (%) |
| м •нҷ•лҸ„ (Accuracy) | (629 + 123) / (629 + 177 + 71 + 123) | 75.20% |
| лҜјк°җлҸ„ (sensitivity) | 123 / (177 + 123) | 41.00% |
| нҠ№мқҙлҸ„ (specificity) | 629 / (629 + 71) | 89.86% |
| мҳӨлҘҳмңЁ (error) | (177 + 71 ) / (629 + 177 + 71 + 123) | 24.80% |
лӢӨмқҢкіј к°ҷмқҙ RлЎң кі„мӮ°н•ҙлҸ„ лҗңлӢӨ.
library("SDMTools")
mat <- confusion.matrix(german$y,german$pred,threshold=0.5)
mat
omission(mat)
sensitivity(mat)
specificity(mat)
prop.correct(mat)
кІ°кіј
> mat
obs
pred 0 1
0 629 177
1 71 123
attr(,"class")
[1] "confusion.matrix"
> omission(mat)
[1] 0.59
> sensitivity(mat)
[1] 0.41
> specificity(mat)
[1] 0.8985714
> prop.correct(mat)
[1] 0.752
>
лӢӨмқҢкіј к°ҷмқҙ н•ҳл©ҙ лӢӨ лӮҳмҳЁлӢӨ.
> accuracy(german$y,german$pred,threshold=0.5)
threshold AUC omission.rate sensitivity specificity prop.correct Kappa
1 0.5 0.6542857 0.59 0.41 0.8985714 0.752 0.3432203
>
к°’м—җ лҢҖн•ң м„ӨлӘ…
- AUC(area under curve): ROC curve м•„лһҳмқҳ л©ҙм Ғ, мҲ«мһҗк°Җ лҶ’мқ„мҲҳлЎқ мўӢлӢӨ.
- omission.rate: the ommission rate as a proportion of true occurrences misidentified given the defined threshold value
- sensitivity: 1мқј нҷ•лҘ , лҜјк°җлҸ„
- specificity: 0мқј нҷ•лҘ , нҠ№мқҙлҸ„
- prop.correct: лӘЁлҚёмқҳ м •нҷ•лҸ„
- kappa statistic
- Fleissмқҳ нҢҗлӢЁкё°мӨҖ..
- мӢ лў°лҸ„ лӮ®лӢӨ: 0.4 лҜёл§Ң
- мӢ лў°лҸ„ мһҲлӢӨ: 0.4 ~ 0.6
- мӢ лў°лҸ„ лҶ’лӢӨ: 0.6 ~ 0.75
- мӢ лў°лҸ„ л§Өмҡ° лҶ’лӢӨ: 0.75 мҙҲкіј
м°ёкі : мөңм Ғ(м •нҷ•лҸ„)мқҳ threshold м°ҫкё°
for (i in seq(0,1,0.01))
{
rs <- c(rs, accuracy(x2$t0,x2$pred,threshold=i)$prop.correct)
}
max_val = max(rs)
for (i in seq(0,1,0.01))
{
tmp <- accuracy(x2$t0,x2$pred,threshold=i)$prop.correct
if ( max_val == tmp )
{
print (paste("threshold = ", i))
}
}
9 ROC(Receiver-Operating Characteristic curve) кіЎм„ #
install.packages("ROCR")
library("ROCR")
cname <- c("x1", "x2", "x3","x4", "x5", "x6", "y")
german = read.table("c:\\data\\german1.txt", col.names = cname)
options(contrasts = c("contr.treatment", "contr.poly"))
german.object <- glm(formula = y ~ x1 + x2 + x3 + x5, family = binomial, data = german)
pred <- predict(german.object, german, type="response")
logodds <- predict(german.object, german)
german <- cbind(german, pred)
german <- cbind(german, logodds)
roc = prediction(german$pred, german$y)
perf = performance(roc, "tpr", "fpr")
#"tpr"мқҖ hit rate, "fpr"мқҖ н—ӣкІҪліҙмңЁ(false alarm rate)
plot(perf)
#plot(perf, print.cutoff.at=seq(0,1,by=0.1))
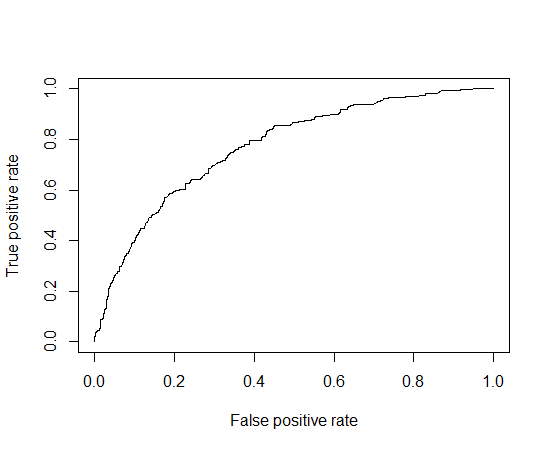
11 кІҖм • #
install.packages("logistf")
library("logistf")
lr2 = logistf(y ~ x1 + x2 + x3 + x5, data = german)
summary(lr2)
кІ°кіј
> summary(lr2)
logistf(formula = y ~ x1 + x2 + x3 + x5, data = german)
Model fitted by Penalized ML
Confidence intervals and p-values by Profile Likelihood
coef se(coef) lower 0.95 upper 0.95 Chisq
(Intercept) 0.64472876 0.474759083 -0.27043182 1.581359116 1.89900357
x1A12 -0.52660853 0.185120846 -0.88988344 -0.166823398 8.25198090
x1A13 -1.04548754 0.334474867 -1.72559520 -0.417128337 11.03319913
x1A14 -1.87794257 0.205266409 -2.28709164 -1.483111213 Inf
x2 0.03387929 0.006308928 0.02165981 0.046313848 29.94733537
x3A31 -0.09194231 0.473477203 -1.01911361 0.824239056 0.03855106
x3A32 -0.92696865 0.369386547 -1.66277580 -0.221680910 6.67102543
x3A33 -0.90779881 0.431809628 -1.76384501 -0.081117723 4.64170188
x3A34 -1.49870282 0.392307541 -2.27839153 -0.748208241 15.50420478
x5 -0.01252255 0.007063063 -0.02651766 0.001115517 3.23324577
p
(Intercept) 1.681899e-01
x1A12 4.070755e-03
x1A13 8.949457e-04
x1A14 0.000000e+00
x2 4.439416e-08
x3A31 8.443407e-01
x3A32 9.799279e-03
x3A33 3.120404e-02
x3A34 8.232193e-05
x5 7.215754e-02
Likelihood ratio test=200.5529 on 9 df, p=0, n=1000
Wald test = 152.8214 on 9 df, p = 0
Covariance-Matrix:
[,1] [,2] [,3] [,4] [,5]
[1,] 0.225396187 -2.116841e-02 -0.0178520944 -1.475598e-02 -1.030299e-03
[2,] -0.021168410 3.426973e-02 0.0158947775 1.663656e-02 -4.993463e-05
[3,] -0.017852094 1.589478e-02 0.1118734364 1.582290e-02 1.008258e-04
[4,] -0.014755985 1.663656e-02 0.0158228986 4.213430e-02 -6.926150e-05
[5,] -0.001030299 -4.993463e-05 0.0001008258 -6.926150e-05 3.980257e-05
[6,] -0.129133562 4.653294e-03 0.0017777035 2.271459e-03 1.822720e-04
[7,] -0.134642541 3.932850e-03 0.0010335670 1.458292e-04 2.134383e-04
[8,] -0.118628622 -2.470301e-03 0.0005935660 -5.484439e-03 -2.417103e-05
[9,] -0.126861265 4.418244e-03 0.0018526164 -2.134237e-03 2.052452e-04
[10,] -0.001743630 7.292805e-05 -0.0000437410 1.938701e-05 -1.300694e-07
[,6] [,7] [,8] [,9] [,10]
[1,] -1.291336e-01 -1.346425e-01 -1.186286e-01 -0.1268612649 -1.743630e-03
[2,] 4.653294e-03 3.932850e-03 -2.470301e-03 0.0044182436 7.292805e-05
[3,] 1.777704e-03 1.033567e-03 5.935660e-04 0.0018526164 -4.374100e-05
[4,] 2.271459e-03 1.458292e-04 -5.484439e-03 -0.0021342366 1.938701e-05
[5,] 1.822720e-04 2.134383e-04 -2.417103e-05 0.0002052452 -1.300694e-07
[6,] 2.241807e-01 1.258971e-01 1.240776e-01 0.1263132224 -8.388392e-05
[7,] 1.258971e-01 1.364464e-01 1.240557e-01 0.1260949748 7.408724e-05
[8,] 1.240776e-01 1.240557e-01 1.864596e-01 0.1247551153 -9.031551e-05
[9,] 1.263132e-01 1.260950e-01 1.247551e-01 0.1539052064 -1.431441e-04
[10,] -8.388392e-05 7.408724e-05 -9.031551e-05 -0.0001431441 4.988686e-05
мқҙлҹ°кұ°лҸ„ мһҲлӢӨ.
install.packages("survey")
library("survey")
regTermTest(german.object, ~x1 + x2 + x3 + x5)
кІ°кіј
> regTermTest(german.object, ~x1 + x2 + x3 + x5)
Wald test for x1 x2 x3 x5
in glm(formula = y ~ x1 + x2 + x3 + x5, family = binomial, data = german)
F = 17.2046 on 9 and 990 df: p= < 2.22e-16
p-valueк°Җ мЎ°л„Ё мһ‘мңјлӢҲк№Ң к·Җл¬ҙк°Җм„Ө м§Җм§Җ лӘ»н•Ё. мҰү, м°Ёмқҙк°Җ мһҲмқҢ.
![[http]](/moniwiki/imgs/http.png) лҚ°мқҙн„°л§ҲмқҙлӢқ(л°©нҶөлҢҖ көҗм ң)(http://press.knou.ac.kr/home/shop/homeBookDetail.do?isbn=9788920006340,)мқҳ мҳҲм ң(
лҚ°мқҙн„°л§ҲмқҙлӢқ(л°©нҶөлҢҖ көҗм ң)(http://press.knou.ac.kr/home/shop/homeBookDetail.do?isbn=9788920006340,)мқҳ мҳҲм ң( german1.txt) нҷңмҡ©
german1.txt) нҷңмҡ©


