[edit]
1 к°ңмҡ” #
мғқмЎҙ 분м„қмқҳ мЈј кҙҖмӢ¬
- мғқмЎҙ н•ЁмҲҳ(survival function)мқҳ м¶”м •
- мғқмЎҙ н•ЁмҲҳ лҳҗлҠ” мң„н—ҳ н•ЁмҲҳ(hazard function)м—җ мҳҒн–Ҙмқ„ мЈјлҠ” кіөліҖлҹү(corvarate) лҳҗлҠ” мҳҲмёЎ ліҖмҲҳлҘј м°ҫм•„лӮҙм–ҙ к·ё м—°кҙҖмқҳ м •лҸ„лҘј кө¬н•ҳкі мһҗ н•Ё.
- мҡ°мӨ‘лҸ„м ҲлӢЁ: tмӢңм җк№Ңм§Җл§Ң мһҗлЈҢмқҳ мғҒнғңлҘј м•Ң мҲҳ мһҲлӢӨ.
- м—°кө¬ мў…лЈҢл“ұмқҳ мқҙмң лЎң tмӢңм җк№Ңм§Җл§Ң мһҗлЈҢлҘј кҙҖмёЎн•Ё. лӘЁл“ мһҗлЈҢк°Җ мҡ°мӨ‘лҸ„м ҲлӢЁмһҗлЈҢ
- лҜёлҰ¬ м •н•ҙлҶ“мқҖ мӮ¬кұҙ л°ңмғқлҘ м—җ лҸ„лӢ¬н•Ё. лӘЁл“ мһҗлЈҢк°Җ мҡ°мӨ‘лҸ„м ҲлӢЁмһҗлЈҢмқҙм§Җл§Ң, м–ём ң кҙҖмёЎмқҙ мў…лЈҢлҗ м§Җ лӘЁлҰ„.
- мһ„мқҳ мҡ°мӨ‘лҸ„м ҲлӢЁмһҗлЈҢ
- н–үм •мғҒ мӨ‘лҸ„м ҲлӢЁ(administrative censoring): м—°кө¬к°Җ мў…лЈҢлҗ л•Ңк№Ңм§Җ мӮ¬к°„мқҙ мқјм–ҙлӮҳм§Җ м•ҠмқҢ.
- 추м Ғ мӢӨнҢЁ(loss to follow up)
- кІҪмҹҒ мң„н—ҳ лӘЁнҳ•(competing risk): лӢӨлҘё мў…лҘҳмқҳ мӮ¬кұҙ л°ңмғқмңјлЎң мқён•ҙ кҙҖмӢ¬мһҲлҠ” мӮ¬кұҙмқҙ мӨ‘лҸ„м ҲлӢЁлҗң кІҪмҡ°
- н–үм •мғҒ мӨ‘лҸ„м ҲлӢЁ(administrative censoring): м—°кө¬к°Җ мў…лЈҢлҗ л•Ңк№Ңм§Җ мӮ¬к°„мқҙ мқјм–ҙлӮҳм§Җ м•ҠмқҢ.
- м—°кө¬ мў…лЈҢл“ұмқҳ мқҙмң лЎң tмӢңм җк№Ңм§Җл§Ң мһҗлЈҢлҘј кҙҖмёЎн•Ё. лӘЁл“ мһҗлЈҢк°Җ мҡ°мӨ‘лҸ„м ҲлӢЁмһҗлЈҢ
[edit]
3 м ҲлӢЁлҗң мһҗлЈҢ #
- мӮҙм•ҳлҠ”м§Җ мЈҪм—ҲлҠ”м§Җ лӘЁлҘҙлҠ” мһҗлЈҢ.
- н•ҳм§Җл§Ң мӮҙм•„мһҲмқ„ лӢ№мӢңк№Ңм§Җмқҳ лҚ°мқҙн„°лҠ” мң нҡЁн•Ё.
- 5л…„ лҸҷм•Ҳ мӮҙм•„мһҲмқҢмқ„ нҷ•мқён–ҲмңјлӮҳ м „нҷ”лІҲнҳёк°Җ л°”лҖҢм–ҙ мғқмЎҙнҷ•мқёмқҙ м–ҙл Өмҡҙ кІҪмҡ°
[edit]
4 мғқмЎҙн•ЁмҲҳS(t), мң„н—ҳн•ЁмҲҳh(t) #
- мғқмЎҙн•ЁмҲҳ - tмӢңм җм—җ мӮҙм•„мһҲмқ„ нҷ•лҘ
- мң„н—ҳн•ЁмҲҳ - tмӢңм җм—җ мӮҙм•„мһҲм§Җл§Ң кі§ мЈҪмқ„ нҷ•лҘ
[edit]
5 мҳҲм ң:мғқмЎҙн•ЁмҲҳ #
normally
- 0=alive
- 1=dead
- Other choices TRUE/FALSE (TRUE = death) or 1/2 (2=death)
- 0=right censored
- 1=event at time
- 2=left censored
- 3=interval censored
library("RODBC")
conn <- odbcConnect("192.168.201.36",uid="id", pwd="pw")
x <- sqlQuery(conn, "select * from ods.dbo.v_h")
#м„ұлі„лі„лЎң м°Ёмқҙк°Җ мһҲлҠ”к°Җ?
out <- survfit(Surv(time, status==1) ~ gender, data=x)
plot(out, lty=1:2, col=c("red", "blue"))
survdiff(Surv(time, status==1) ~ gender, data=x)
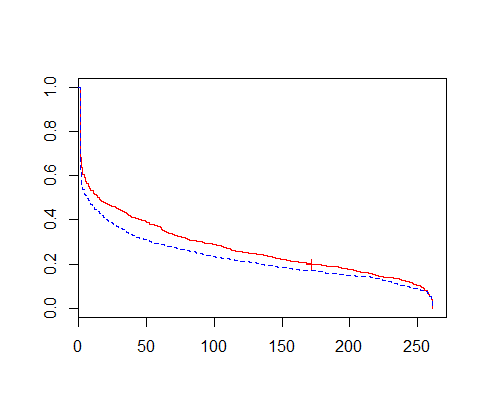
survdiff(Surv(time, status==1) ~ gender, data=x)мқҳ кІ°кіјлҠ” лӢӨмқҢкіј к°ҷлӢӨ. survdiffлҠ” мғқмЎҙн•ЁмҲҳлҘј м–»лҠ”лӢӨ.
> survdiff(Surv(time, status==1) ~ gender, data=x)
Call:
survdiff(formula = Surv(time, status == 1) ~ gender, data = x)
N Observed Expected (O-E)^2/E (O-E)^2/V
gender=0 881 804 852 2.70 6.14
gender=1 1142 1054 1006 2.28 6.14
Chisq= 6.1 on 1 degrees of freedom, p= 0.0132
p-valueк°Җ 0.0132лЎң мң мқҳмҲҳмӨҖ 0.05м—җм„ң к·Җл¬ҙк°Җм„Ө кё°к°Ғ, лҢҖлҰҪк°Җм„Ө мұ„нғқ. мҰү, м„ұлі„лі„лЎң м°Ёмқҙк°Җ мһҲмқҢмқ„ нҷ•мқён• мҲҳ мһҲлӢӨ. summary(out)мқҳ кІ°кіјлҘј ліҙмһҗ.
> summary(out)
Call: survfit(formula = Surv(time, status == 1) ~ gender, data = x)
gender=0
time n.risk n.event survival std.err lower 95% CI upper 95% CI
1 881 281 0.6810 0.01570 0.6510 0.7125
2 525 34 0.6369 0.01641 0.6056 0.6699
3 491 25 0.6045 0.01680 0.5725 0.6384
4 466 9 0.5928 0.01692 0.5606 0.6269
5 457 13 0.5760 0.01708 0.5435 0.6104
6 444 10 0.5630 0.01718 0.5303 0.5977
7 434 9 0.5513 0.01726 0.5185 0.5862
8 425 8 0.5409 0.01732 0.5080 0.5760
9 417 5 0.5345 0.01735 0.5015 0.5696
10 412 2 0.5319 0.01736 0.4989 0.5670
11 410 8 0.5215 0.01741 0.4885 0.5567
12 402 5 0.5150 0.01743 0.4819 0.5503
13 397 4 0.5098 0.01745 0.4767 0.5452
14 393 5 0.5033 0.01747 0.4702 0.5387
15 388 8 0.4929 0.01749 0.4598 0.5284
16 380 3 0.4891 0.01749 0.4559 0.5246
17 377 4 0.4839 0.01750 0.4508 0.5194
18 373 4 0.4787 0.01750 0.4456 0.5142
19 369 1 0.4774 0.01750 0.4443 0.5129
20 368 2 0.4748 0.01750 0.4417 0.5104
21 366 2 0.4722 0.01750 0.4391 0.5078
22 364 2 0.4696 0.01750 0.4365 0.5052
23 362 3 0.4657 0.01750 0.4326 0.5013
24 359 3 0.4618 0.01750 0.4288 0.4974
.
.
.
gender=1
time n.risk n.event survival std.err lower 95% CI upper 95% CI
1 1142 437 0.6173 0.01438 0.5898 0.6462
2 618 57 0.5604 0.01490 0.5319 0.5904
3 561 24 0.5364 0.01505 0.5077 0.5667
4 537 20 0.5164 0.01514 0.4876 0.5470
5 517 7 0.5095 0.01516 0.4806 0.5400
6 510 10 0.4995 0.01519 0.4706 0.5301
7 500 11 0.4885 0.01521 0.4596 0.5192
8 489 6 0.4825 0.01522 0.4536 0.5133
9 483 11 0.4715 0.01523 0.4426 0.5023
10 472 9 0.4625 0.01523 0.4336 0.4933
11 463 4 0.4585 0.01523 0.4296 0.4894
12 459 11 0.4475 0.01522 0.4187 0.4784
13 448 4 0.4435 0.01522 0.4147 0.4744
14 444 7 0.4365 0.01520 0.4077 0.4674
15 437 6 0.4305 0.01519 0.4018 0.4614
16 431 7 0.4235 0.01517 0.3948 0.4544
17 424 5 0.4186 0.01516 0.3899 0.4493
18 419 9 0.4096 0.01512 0.3810 0.4403
19 410 2 0.4076 0.01512 0.3790 0.4383
20 408 5 0.4026 0.01510 0.3740 0.4333
21 403 5 0.3976 0.01507 0.3691 0.4282
22 398 4 0.3936 0.01505 0.3652 0.4242
.
.
.
к°Ғ мӢңм җлі„лЎң мЈҪмқ„ нҷ•лҘ мқҙ лӮҳмҳЁлӢӨ. м—¬мһҗлҠ” 5мқјм°Ёк№Ңм§Җ мӮҙм•„мһҲмқ„ нҷ•лҘ мқҙ 0.5760мқҙкі , лӮЁмһҗлҠ” 0.5095лӢӨ. [edit]
6 мҳҲм ң:мң„н—ҳн•ЁмҲҳ #
cox regressionмқ„ м“ҙлӢӨ.
> summary(coxph(Surv(time, status==1) ~ gender, data=x))
Call:
coxph(formula = Surv(time, status == 1) ~ gender, data = x)
n= 607, number of events= 605
coef exp(coef) se(coef) z Pr(>|z|)
gender 0.10346 1.10900 0.08214 1.26 0.208
exp(coef) exp(-coef) lower .95 upper .95
gender 1.109 0.9017 0.9441 1.303
Concordance= 0.527 (se = 0.033 )
Rsquare= 0.003 (max possible= 1 )
Likelihood ratio test= 1.59 on 1 df, p=0.2069
Wald test = 1.59 on 1 df, p=0.2078
Score (logrank) test = 1.59 on 1 df, p=0.2076
м—¬мһҗм—җ 비н•ҙ лӮЁмһҗмқҳ мҲңк°„мӮ¬л§қмң„н—ҳ(hazard)к°Җ 1.10900л°° лҶ’лӢӨ. p-valueк°Җ 0.208лЎң мң мқҳн•ҳм§ҖлҠ” м•ҠлӢӨ.[edit]
7 мҳҲм ң #
out2 <- survfit(Surv(suvival_time, status==1) ~ mm, data=m)
summary(out2)
col <- colors()[c(26,31,33,51,76,153)]
plot(out2, lty=1:2, col=col)
legend("topright", levels(m$mm), col=col, text.col=col)
col <- colors()[c(26,31,33,36,50,62,70,128,142,258,367,477,275)]
plot(fit2, col=col, lwd=1)
legend("topright", levels(sur$к°Җмһ…мӣ”), col=col, text.col=col)
[edit]
8 interval #
library(survival)
x <- df[df$genre == "нҚјмҰҗ",]
out <- survfit(Surv(begin_dt, end_dt, event, type="interval") ~ nation, data=df)
col <- colors()[c(26,31,33,36,50,62,70,128,142,258,367,477,275)]
plot(out, col=col)
legend("topright", levels(df$nation), col=col, text.col=col)
s <- summary(out)
data.frame(nation=gsub("nation=", "", s$strata), time=s$time, n.risk=s$n.risk, n.event=s$n.event, survival=s$surv)
[edit]
9 AUC(area under the curve) #
#AUC
install.packages("pracma")
require(pracma)
out <- survfit(Surv(time=diff, event=event, type="right") ~ nation, data=x)
rs <- data.frame(time=summary(out)$time, surv=summary(out)$surv, nation=gsub("nation=", "",summary(out)$strata))
trapz(rs[rs$nation=="мӨ‘көӯ",]$time, rs[rs$nation=="мӨ‘көӯ",]$surv)
[edit]
10 ctree #
library(party)
library(survival)
data("GBSG2", package = "TH.data")
fit <- ctree(Surv(time, cens) ~ ., data = GBSG2)
plot(fit)
[edit]
11 survfit кІ°кіјлҘј data.frame мңјлЎң #
https://github.com/kmiddleton/rexamples/blob/master/qplot_survival.R
createSurvivalFrame <- function(f.survfit){
# initialise frame variable
f.frame <- NULL
# check if more then one strata
if(length(names(f.survfit$strata)) == 0){
# create data.frame with data from survfit
f.frame <- data.frame(time=f.survfit$time,
n.risk=f.survfit$n.risk,
n.event=f.survfit$n.event,
n.censor = f.survfit$n.censor,
surv=f.survfit$surv,
upper=f.survfit$upper,
lower=f.survfit$lower)
# create first two rows (start at 1)
f.start <- data.frame(time=c(0, f.frame$time[1]),
n.risk=c(f.survfit$n, f.survfit$n),
n.event=c(0,0),
n.censor=c(0,0),
surv=c(1,1),
upper=c(1,1),
lower=c(1,1))
# add first row to dataset
f.frame <- rbind(f.start, f.frame)
# remove temporary data
rm(f.start)
}
else {
# create vector for strata identification
f.strata <- NULL
for(f.i in 1:length(f.survfit$strata)){
# add vector for one strata according to number of rows of strata
f.strata <- c(f.strata, rep(names(f.survfit$strata)[f.i], f.survfit$strata[f.i]))
}
# create data.frame with data from survfit (create column for strata)
f.frame <- data.frame(time=f.survfit$time, n.risk=f.survfit$n.risk, n.event=f.survfit$n.event, n.censor = f.survfit$n.censor, surv=f.survfit$surv, upper=f.survfit$upper, lower=f.survfit$lower, strata=factor(f.strata))
# remove temporary data
rm(f.strata)
# create first two rows (start at 1) for each strata
for(f.i in 1:length(f.survfit$strata)){
# take only subset for this strata from data
f.subset <- subset(f.frame, strata==names(f.survfit$strata)[f.i])
# create first two rows (time: 0, time of first event)
f.start <- data.frame(time=c(0, f.subset$time[1]), n.risk=rep(f.survfit[f.i]$n, 2), n.event=c(0,0), n.censor=c(0,0), surv=c(1,1), upper=c(1,1), lower=c(1,1), strata=rep(names(f.survfit$strata)[f.i],2))
# add first two rows to dataset
f.frame <- rbind(f.start, f.frame)
# remove temporary data
rm(f.start, f.subset)
}
# reorder data
f.frame <- f.frame[order(f.frame$strata, f.frame$time), ]
# rename row.names
rownames(f.frame) <- NULL
}
# return frame
return(f.frame)
}
[edit]
12 м№ҙн”ҢлһҖ-л§Ҳмқҙм–ҙ л°©лІ• #
мң„ м—‘м…Җм—җм„ң лҲ„м ҒмғқмЎҙмңЁмқҳ н•©мқҖ restricted mean survival time (RMST) лӢӨ.
RмҪ”л“ңлЎң н•ҳл©ҙ..
RмҪ”л“ңлЎң н•ҳл©ҙ..
library("survival")
time <- c(3,2,2,1,0,0,0)
status <- c(1,1,1,1,1,1,1)
fit <- survfit(Surv(time,status) ~ 1)
print(fit, print.rmean=TRUE)
plot(fit)
rmst <- sum(fit$surv)
[edit]
13 м°ёкі мһҗлЈҢ #
- https://rviews.rstudio.com/2017/09/25/survival-analysis-with-r/
- http://www.r-statistics.com/2013/07/creating-good-looking-survival-curves-the-ggsurv-function/ --> ggsurv мқҙмҒҳкІҢ к·ёл ӨмӨҢ.
- http://www.ats.ucla.edu/stat/r/examples/asa/default.htm
 кі к°қ세분нҷ”м—җ_кё°л°ҳн•ң_мғқмЎҙ분м„қмқ„_нҷңмҡ©н•ң_кі к°қмҲҳлӘ…мҳҲмёЎлӘЁлҚё
кі к°қ세분нҷ”м—җ_кё°л°ҳн•ң_мғқмЎҙ분м„қмқ„_нҷңмҡ©н•ң_кі к°қмҲҳлӘ…мҳҲмёЎлӘЁлҚё
лӯҳ м°ҫлӢӨк°Җ ліҙл©ҙ лӢӨмӢң мқҙ мӮ¬мқҙнҠёлЎң мҳӨлҠ” кІҪмҡ°к°Җ л§ҺмҠөлӢҲлӢӨ. мўӢмқҖ мһҗлЈҢ кіөмң н•ҙ мЈјм…”м…” к°җмӮ¬н•©лӢҲлӢӨ. -- мһҘмҡҙнҳё 2014-11-11 10:16:39
г…Ӣ к·ёлҹ°к°Җмҡ”~ -- мқҙмһ¬н•ҷ 2014-11-12 16:14:52
п»ҝ
![[http]](/moniwiki/imgs/http.png)