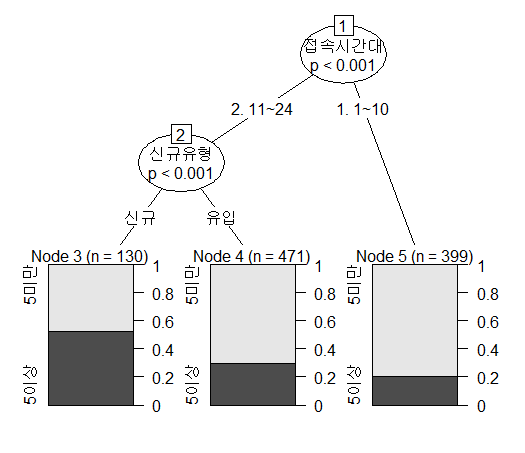
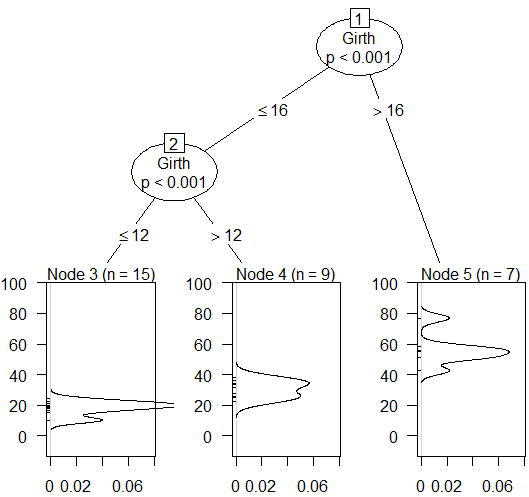
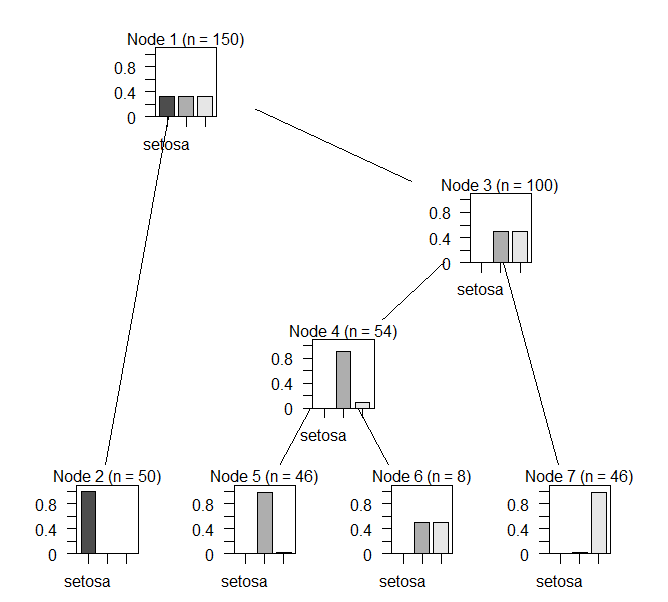
--
http://stackoverflow.com/questions/11961923/rotate-classification-tree-terminal-barplot-axis-r
# Note inclusion of horiz = FALSE
alt_node_barplot <- function (ctreeobj, col = "black", fill = NULL, beside = NULL,
ymax = NULL, ylines = NULL, widths = 1, gap = NULL, reverse = NULL,
id = TRUE, horiz = FALSE)
{
getMaxPred <- function(x) {
mp <- max(x$prediction)
mpl <- ifelse(x$terminal, 0, getMaxPred(x$left))
mpr <- ifelse(x$terminal, 0, getMaxPred(x$right))
return(max(c(mp, mpl, mpr)))
}
y <- response(ctreeobj)[[1]]
if (is.factor(y) || class(y) == "was_ordered") {
ylevels <- levels(y)
if (is.null(beside))
beside <- if (length(ylevels) < 3)
FALSE
else TRUE
if (is.null(ymax))
ymax <- if (beside)
1.1
else 1
if (is.null(gap))
gap <- if (beside)
0.1
else 0
}
else {
if (is.null(beside))
beside <- FALSE
if (is.null(ymax))
ymax <- getMaxPred(ctreeobj@tree) * 1.1
ylevels <- seq(along = ctreeobj@tree$prediction)
if (length(ylevels) < 2)
ylevels <- ""
if (is.null(gap))
gap <- 1
}
if (is.null(reverse))
reverse <- !beside
if (is.null(fill))
fill <- gray.colors(length(ylevels))
if (is.null(ylines))
ylines <- if (beside)
c(3, 2)
else c(1.5, 2.5)
# My edit do not work if beside is not true
#################################################
if(!beside) horiz = FALSE
#################################################
rval <- function(node) {
pred <- node$prediction
if (reverse) {
pred <- rev(pred)
ylevels <- rev(ylevels)
}
np <- length(pred)
nc <- if (beside)
np
else 1
fill <- rep(fill, length.out = np)
widths <- rep(widths, length.out = nc)
col <- rep(col, length.out = nc)
ylines <- rep(ylines, length.out = 2)
gap <- gap * sum(widths)
#######################################################
if (!horiz){
yscale <- c(0, ymax)
xscale <- c(0, sum(widths) + (nc + 1) * gap)
} else {
xscale <- c(0, ymax)
yscale <- c(0, sum(widths) + (nc + 1) * gap)
}
#######################################################
top_vp <- viewport(layout = grid.layout(nrow = 2, ncol = 3,
widths = unit(c(ylines[1], 1, ylines[2]), c("lines",
"null", "lines")), heights = unit(c(1, 1), c("lines",
"null"))), width = unit(1, "npc"), height = unit(1,
"npc") - unit(2, "lines"), name = paste("node_barplot",
node$nodeID, sep = ""))
pushViewport(top_vp)
grid.rect(gp = gpar(fill = "white", col = 0))
top <- viewport(layout.pos.col = 2, layout.pos.row = 1)
pushViewport(top)
mainlab <- paste(ifelse(id, paste("Node", node$nodeID,
"(n = "), "n = "), sum(node$weights), ifelse(id,
")", ""), sep = "")
grid.text(mainlab)
popViewport()
plot <- viewport(layout.pos.col = 2, layout.pos.row = 2,
xscale = xscale, yscale = yscale, name = paste("node_barplot",
node$nodeID, "plot", sep = ""))
pushViewport(plot)
if (beside) {
#############################################################
if(!horiz){
xcenter <- cumsum(widths + gap) - widths/2
for (i in 1:np) {
grid.rect(x = xcenter[i], y = 0, height = pred[i],
width = widths[i], just = c("center", "bottom"),
default.units = "native", gp = gpar(col = col[i],
fill = fill[i]))
}
if (length(xcenter) > 1)
grid.xaxis(at = xcenter, label = FALSE)
grid.text(ylevels, x = xcenter, y = unit(-1, "lines"),
just = c("center", "top"), default.units = "native",
check.overlap = TRUE)
grid.yaxis()
} else {
ycenter <- cumsum(widths + gap) - widths/2
for (i in 1:np) {
grid.rect(y = ycenter[i], x = 0, width = pred[i],
height = widths[i], just = c("left", "center"),
default.units = "native", gp = gpar(col = col[i],
fill = fill[i]))
}
if (length(ycenter) > 1)
grid.yaxis(at = ycenter, label = FALSE)
grid.text(ylevels, y = ycenter, x = unit(-1, "lines"),
just = c("right", "center"), default.units = "native",
check.overlap = TRUE)
grid.xaxis()
}
#############################################################
}
else {
ycenter <- cumsum(pred) - pred
for (i in 1:np) {
grid.rect(x = xscale[2]/2, y = ycenter[i], height = min(pred[i],
ymax - ycenter[i]), width = widths[1], just = c("center",
"bottom"), default.units = "native", gp = gpar(col = col[i],
fill = fill[i]))
}
if (np > 1) {
grid.text(ylevels[1], x = unit(-1, "lines"),
y = 0, just = c("left", "center"), rot = 90,
default.units = "native", check.overlap = TRUE)
grid.text(ylevels[np], x = unit(-1, "lines"),
y = ymax, just = c("right", "center"), rot = 90,
default.units = "native", check.overlap = TRUE)
}
if (np > 2) {
grid.text(ylevels[-c(1, np)], x = unit(-1, "lines"),
y = ycenter[-c(1, np)], just = "center", rot = 90,
default.units = "native", check.overlap = TRUE)
}
grid.yaxis(main = FALSE)
}
grid.rect(gp = gpar(fill = "transparent"))
upViewport(2)
}
return(rval)
}