[edit]
3 ņśłņĀ£ #
#install.packages("tree")
library(tree)
ir.tr <- tree(Species ~., iris)
ir.tr
ir.tr1 <- snip.tree(ir.tr, nodes = c(12, 7))
summary(ir.tr1)
par(pty = "s")
plot(iris[, 3],iris[, 4], type="n",
xlab="petal length", ylab="petal width")
text(iris[, 3], iris[, 4], c("s", "c", "v")[iris[, 5]])
partition.tree(ir.tr1, add = TRUE, cex = 1.5)
# 1D example
ir.tr <- tree(Petal.Width ~ Petal.Length, iris)
plot(iris[,3], iris[,4], type="n", xlab="Length", ylab="Width")
partition.tree(ir.tr, add = TRUE, cex = 1.5)
partition.tree()ļź╝ ņé¼ņÜ®ĒĢśļ®┤ ņĢäļלņÖĆ Ļ░ÖņØĆ ĻĘĖļ”╝ņØä ļ│╝ ņłś ņ׳ļŗż. 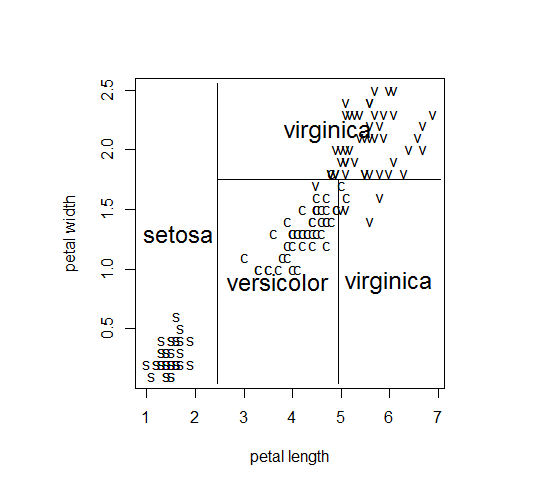

![[http]](/moniwiki/imgs/http.png)