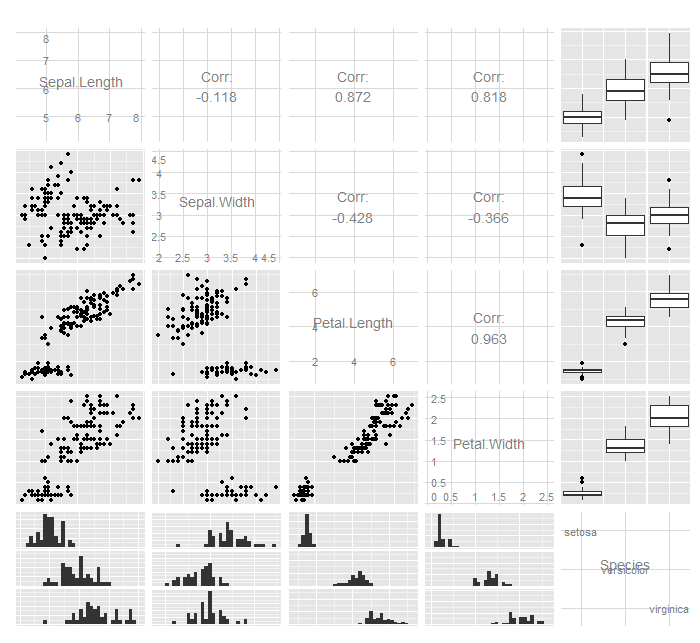
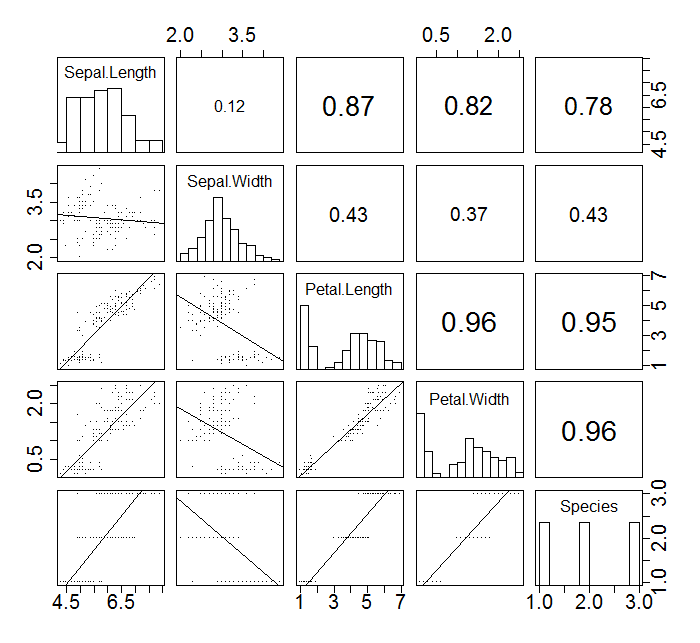
[edit]
1 3d surface #
https://stackoverflow.com/questions/41700400/smoothing-3d-plot-in-r
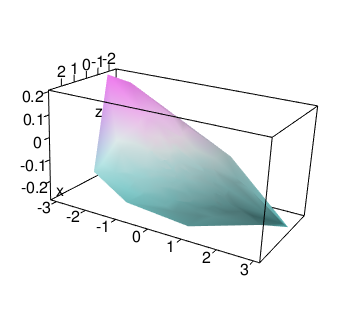
library(mgcv)
x<- rnorm(200)
y<- rnorm(200)
z<-rnorm(200)
tab<-data.frame(x,y,z)
tab
#surface wireframe:
mod <- gam(z ~ te(x, y), data = tab)
library(rgl)
library(deldir)
zfit <- fitted(mod)
col <- cm.colors(20)[1 +
round(19*(zfit - min(zfit))/diff(range(zfit)))]
persp3d(deldir(x, y, z = zfit), col = col)
aspect3d(1, 2, 1)

![[https]](/moniwiki/imgs/https.png)