![[https]](/moniwiki/imgs/https.png) June 2017 New Package Picks(https://www.r-bloggers.com/june-2017-new-package-picks/)
June 2017 New Package Picks(https://www.r-bloggers.com/june-2017-new-package-picks/)
![[https]](/moniwiki/imgs/https.png) June 2017 New Package Picks(https://www.r-bloggers.com/june-2017-new-package-picks/)
June 2017 New Package Picks(https://www.r-bloggers.com/june-2017-new-package-picks/)
eom <- function(date) {
# date character string containing POSIXct date
date.lt <- as.POSIXlt(date) # add a month, then subtract a day:
mon <- date.lt$mon + 2
year <- date.lt$year
year <- year + as.integer(mon==13) # if month was December add a year
mon[mon==13] <- 1
iso = ISOdate(1900+year, mon, 1, hour=0, tz=attr(date,"tz"))
result = as.POSIXct(iso) - 86400 # subtract one day
result + (as.POSIXlt(iso)$isdst - as.POSIXlt(result)$isdst)*3600
}
#eom(as.POSIXct("2015-02-01"))
right <- function (string, char){
substr(string,nchar(string)-(char-1),nchar(string))
}
left <- function (string,char){
substr(string,1,char)
}
as.lm <- function(object, ...) {
if (!inherits(object, "nls")) {
w <- paste("expected object of class nls but got object of class:",
paste(class(object), collapse = " "))
warning(w)
}
gradient <- object$m$gradient()
if (is.null(colnames(gradient))) {
colnames(gradient) <- names(object$m$getPars())
}
response.name <- if (length(formula(object)) == 2) "0" else
as.character(formula(object)[[2]])
lhs <- object$m$lhs()
L <- data.frame(lhs, gradient)
names(L)[1] <- response.name
fo <- sprintf("%s ~ %s - 1", response.name,
paste(colnames(gradient), collapse = "+"))
fo <- as.formula(fo, env = as.proto.list(L))
do.call("lm", list(fo, offset = substitute(fitted(object))))
}
crossjoin <- function(...,FUN='data.frame') {
args <- list(...)
n1 <- names(args)
n2 <- sapply(match.call()[1+1:length(args)], as.character)
nn <- if (is.null(n1)) n2 else ifelse(n1!='',n1,n2)
dims <- sapply(args,length)
dimtot <- prod(dims)
reps <- rev(cumprod(c(1,rev(dims))))[-1]
cols <- lapply(1:length(dims), function(j)
args[[j]][1+((1:dimtot-1) %/% reps[j]) %% dims[j]])
names(cols) <- nn
do.call(match.fun(FUN),cols)
}
add.error.bars <- function(X,Y,SE,w,col=1){
X0 = X; Y0 = (Y-SE); X1 =X; Y1 = (Y+SE);
arrows(X0, Y0, X1, Y1, code=3,angle=90,length=w,col=col);
}
plot(x,y, ylim=c(lwr,upr))
add.error.bars(last(x), last(val), pred$se, 0.1)
expr <- function(model){
f <- formula(model)
rhs <- unlist(strsplit(as.character(f), "~"))[2]
lhs <- unlist(strsplit(as.character(f), "~"))[3]
for(v in names(coef(model))){
lhs <- gsub(v, paste0(coef(model)[v], "*", v), lhs)
}
result <- paste(rhs, "~", lhs)
return (result)
}
model <- lm(Volume ~ Girth + Height, data=trees)
Girth <- trees$Girth
Height <- trees$Height
txt <- unlist(strsplit(as.character(expr(model)), "~"))[2]
eval(parse( text=txt))
library("pryr")
eval(parse(text=paste0("ast(", txt, ")")))
out <- capture.output(eval(parse(text=paste0("ast(", txt, ")"))))
out
gsub("([\\])-", "", out)
> eval(parse( text=txt))
[1] 62.82532 62.54151 62.80464 73.86177 77.85667 79.00599 74.18035 77.23361 79.40068
[10] 78.17524 80.00306 79.45612 79.45612 78.49381 81.94177 85.83986 89.57163 91.79414
[19] 88.58864 86.68469 92.37584 93.99598 93.37292 99.75666 102.86536 108.93053 110.21141
[28] 111.41617 111.88699 111.88699 126.50296
> eval(parse(text=paste0("ast(", txt, ")")))
\- ()
\- `+
\- ()
\- `*
\- 4.70816050301751
\- `Girth
\- ()
\- `*
\- 0.339251234244701
\- `Height
> out <- capture.output(eval(parse(text=paste0("ast(", txt, ")"))))
> out
[1] "\\- ()" " \\- `+" " \\- ()"
[4] " \\- `*" " \\- 4.70816050301751" " \\- `Girth"
[7] " \\- ()" " \\- `*" " \\- 0.339251234244701"
[10] " \\- `Height "
> gsub("([\\])-", "", out)
[1] " ()" " `+" " ()"
[4] " `*" " 4.70816050301751" " `Girth"
[7] " ()" " `*" " 0.339251234244701"
[10] " `Height "
lagpad <- function(x, k) {
if (!is.vector(x))
stop('x must be a vector')
if (!is.numeric(x))
stop('x must be numeric')
if (!is.numeric(k))
stop('k must be numeric')
if (1 != length(k))
stop('k must be a single number')
c(rep(NA, k), x)[1 : length(x)]
}
library(DataCombine) slide(data.frame(DAU), Var = "DAU", slideBy = -3)
shannon.entropy <- function(p)
{
if (min(p) < 0 || sum(p) <= 0)
return(NA)
p.norm <- p[p>0]/sum(p)
-sum(log2(p.norm)*p.norm)
}
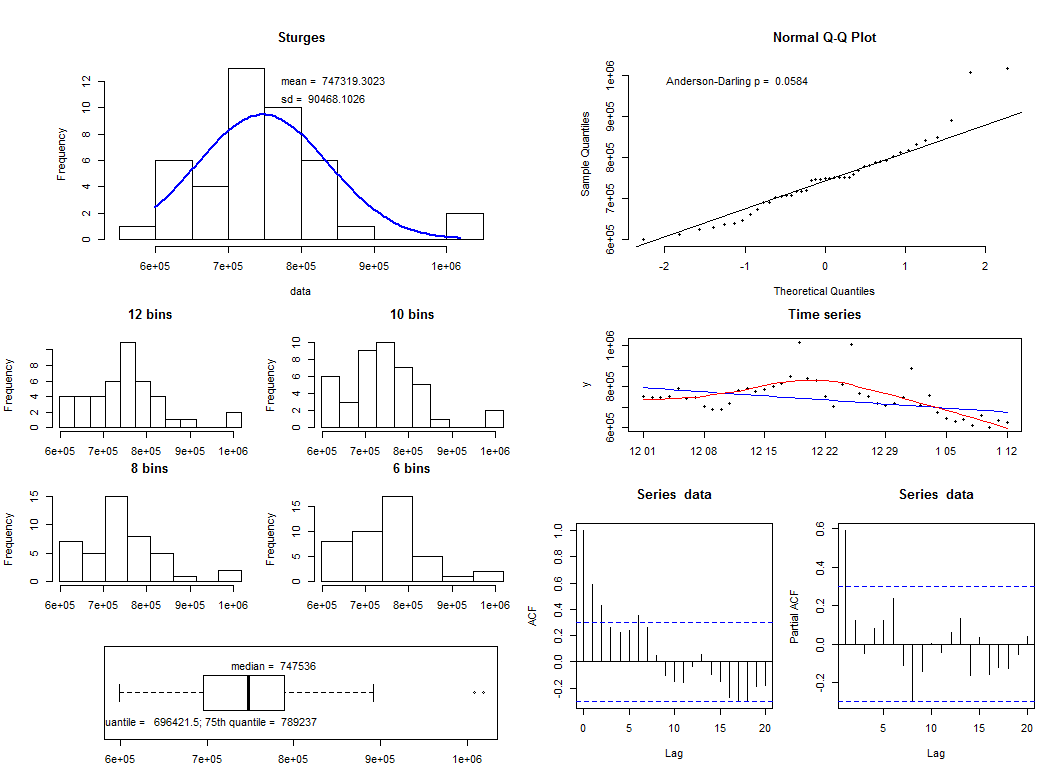
summary.plot <- function(data, xlabels){
require(nortest)
require(MASS)
library(tree)
library(mgcv)
# first job is to save the graphics parameters currently used
def.par <- par(no.readonly = TRUE)
par("plt" = c(.2,.95,.2,.8))
layout( matrix(c(1,1,2,2,1,1,2,2,4,5,8,8,6,7,9,10,3,3,9,10), 5, 4, byrow = TRUE))
#histogram on the top left
h <- hist(data, breaks = "Sturges", plot = FALSE)
xfit<-seq(min(data),max(data),length=100)
yfit<-yfit<-dnorm(xfit,mean=mean(data),sd=sd(data))
yfit <- yfit*diff(h$mids[1:2])*length(data)
plot (h, axes = TRUE, main = "Sturges")
lines(xfit, yfit, col="blue", lwd=2)
leg1 <- paste("mean = ", round(mean(data), digits = 4))
leg2 <- paste("sd = ", round(sd(data),digits = 4))
legend(x = "topright", c(leg1,leg2), bty = "n")
## normal qq plot
qqnorm(data, bty = "n", pch = 20)
qqline(data)
p <- ad.test(data)
leg <- paste("Anderson-Darling p = ", round(as.numeric(p[2]), digits = 4))
legend(x = "topleft", leg, bty = "n")
## boxplot (bottom left)
boxplot(data, horizontal = TRUE)
leg1 <- paste("median = ", round(median(data), digits = 4))
lq <- quantile(data, 0.25)
leg2 <- paste("q1 = ", round(lq,digits = 4))
uq <- quantile(data, 0.75)
leg3 <- paste("q3 = ", round(uq,digits = 4))
legend(x = "top", leg1, bty = "n")
legend(x = "bottom", paste(leg2, leg3, sep = "; "), bty = "n", lty = c(1,2))
## the various histograms with different bins
h2 <- hist(data, breaks = (0:12 * (max(data) - min (data))/12)+min(data), plot = FALSE)
plot (h2, axes = TRUE, main = "12 bins")
h3 <- hist(data, breaks = (0:10 * (max(data) - min (data))/10)+min(data), plot = FALSE)
plot (h3, axes = TRUE, main = "10 bins")
h4 <- hist(data, breaks = (0:8 * (max(data) - min (data))/8)+min(data), plot = FALSE)
plot (h4, axes = TRUE, main = "8 bins")
h5 <- hist(data, breaks = (0:6 * (max(data) - min (data))/6)+min(data), plot = FALSE)
plot (h5, axes = TRUE,main = "6 bins")
## the time series, ACF and PACF
#plot (data, main = "Time series", pch = 20)
x <- xlabels
y <- data
#fit1 <- rlm(y~x)
fit1 <- lqs(y~x, method="lts", quantile=8)
pred <- predict(fit1, data.frame(x=x))
lwr <- pred - (IQR(pred)/1.35)*3
upr <- pred + (IQR(pred)/1.35)*3
ylim <- c(min(y)*0.9, max(y)*1.1)
if (min(lwr) < min(y)){
ylim <- c(min(lwr)*0.9, max(y)*1.1)
}
plot(x,y, main="Time series", pch = 20, type="l", ylim=ylim)
points(x,y, main="Time series", pch = 20)
lines(x, pred, col="blue")
lines(x, lwr, col="blue", lty=2)
lines(x, upr, col="blue", lty=2)
#lines(x, predict(fit1, data.frame(x=x)), col="blue")
x <- 1:length(y)
fit2 <- gam(y~s(x))
tree.model <- tree(data.frame(y, x))
pred1 <- predict(tree.model, list(x=x))
x <- xlabels
lines(x, pred1, col="red")
acf(data, lag.max = 20)
pacf(data, lag.max = 20)
## reset the graphics display to default
par(def.par)
}
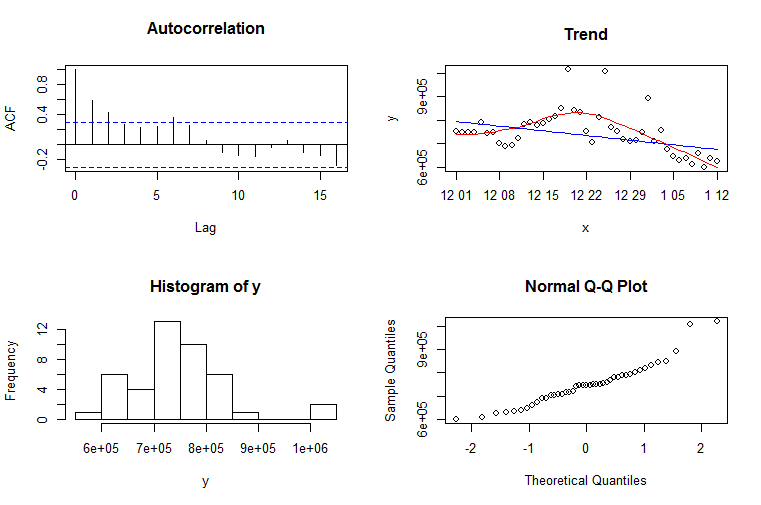
four.plot <- function(y, xlabels){
par(mfrow=c(2,2))
acf(y, main="Autocorrelation")
x <- xlabels
plot(x,y, main="Trend")
fit1 <- rlm(y~x)
lines(x, predict(fit1, data.frame(x=x)), col="blue")
x <- 1:length(y)
fit2 <- loess(y~x)
pred <- predict(fit2, data.frame(x=x))
x <- xlabels
lines(x, pred, col="red")
hist(y)
qqnorm(y)
par(mfrow=c(1,1))
}
robustz <- function(x){
sigma <- IQR(x)/1.35
if(sigma == 0) sigma <- sd(x)
if(sigma == 0) sigma <- 0.0001
z <- (x - median(x)) / sigma
return(z)
}