[edit]
2 мҳҲм ңлҚ°мқҙн„° #
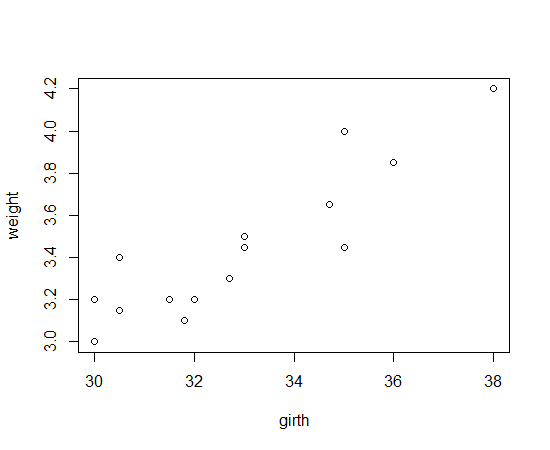
н—ҲлҰ¬л‘ҳл Ҳ(girth)мҷҖ лӘёл¬ҙкІҢ(weigth)
tmp <- textConnection( "girth weight 35.00 3.45 32.00 3.20 30.00 3.00 31.50 3.20 32.70 3.30 30.00 3.20 36.00 3.85 30.50 3.15 34.70 3.65 30.50 3.40 33.00 3.50 35.00 4.00 31.80 3.10 38.00 4.20 33.00 3.45") x <- read.table(tmp, header=TRUE) close.connection(tmp) plot(x)
мӮ°м җлҸ„

![[http]](/moniwiki/imgs/http.png)
